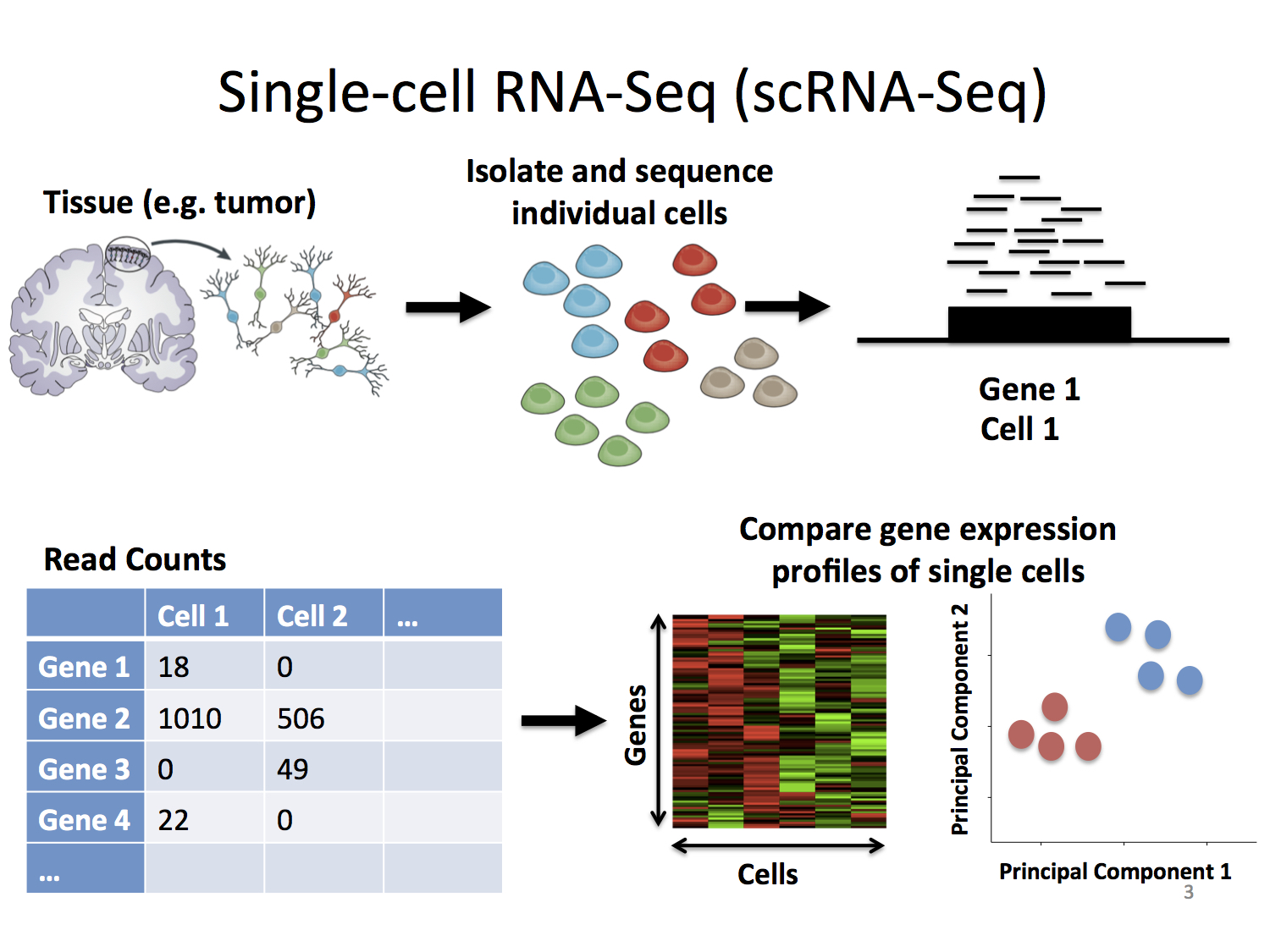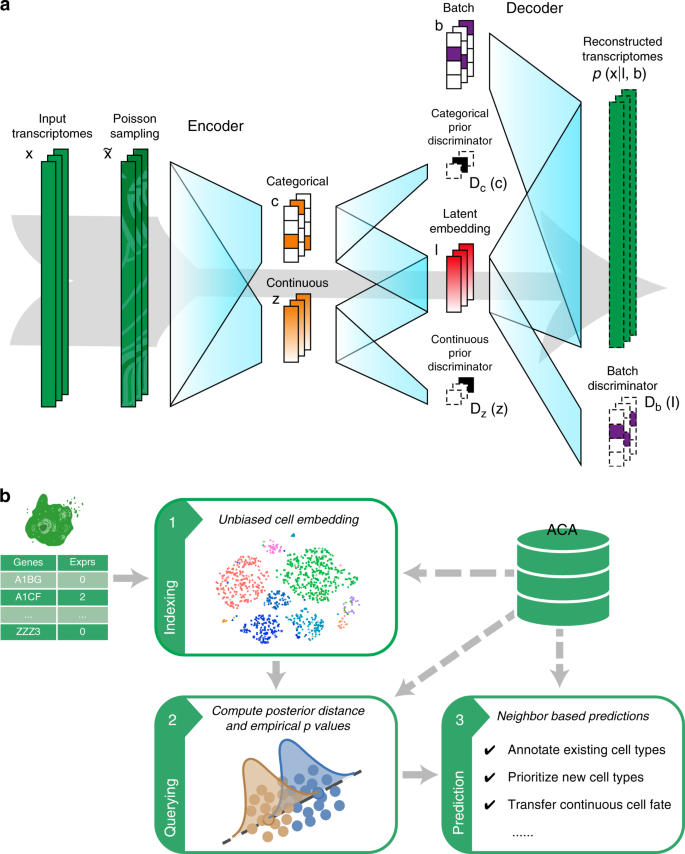
Searching large-scale scRNA-seq databases via unbiased cell embedding with Cell BLAST | Nature Communications
scPipe: A flexible R/Bioconductor preprocessing pipeline for single-cell RNA-sequencing data | PLOS Computational Biology
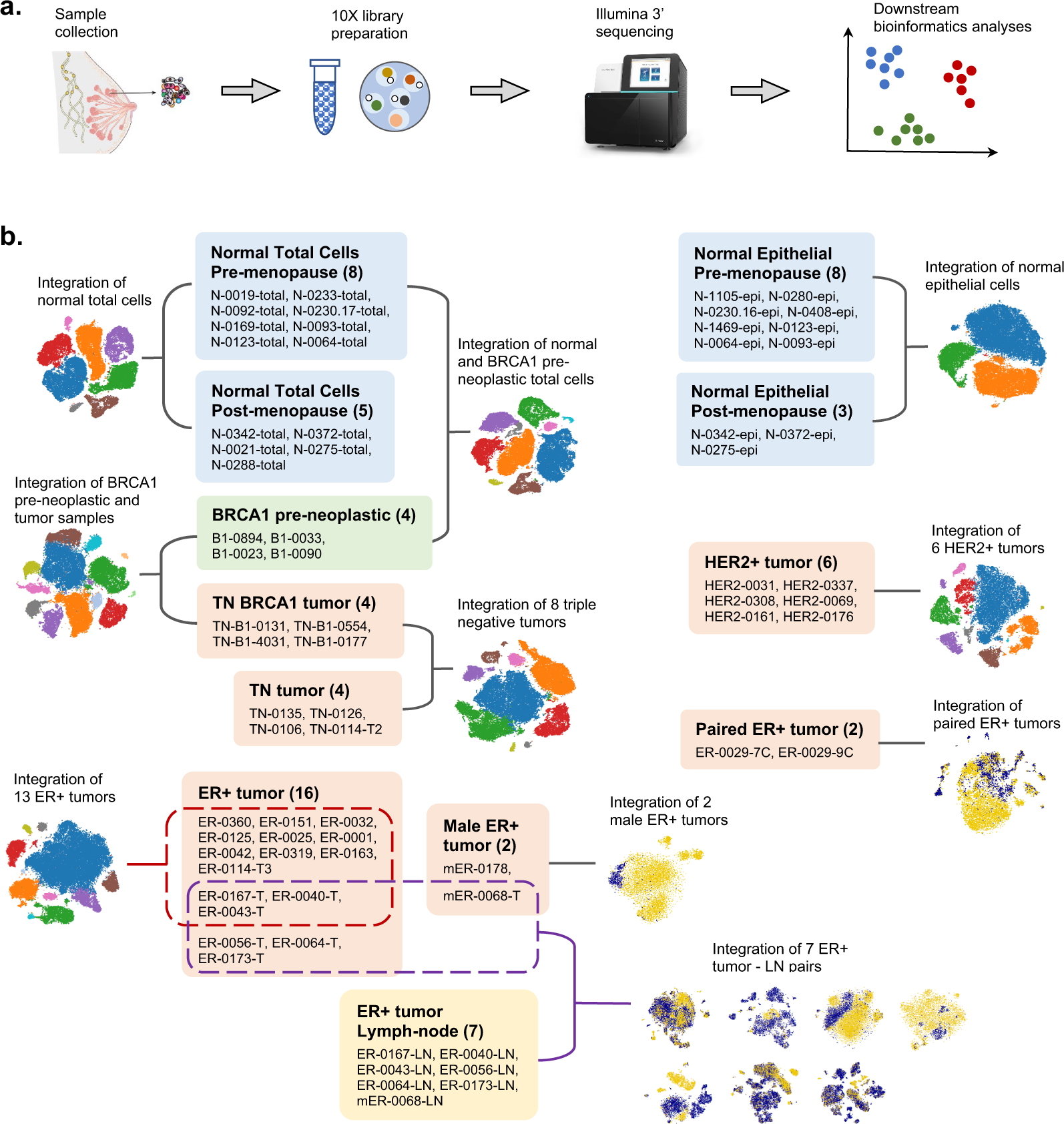
R code and downstream analysis objects for the scRNA-seq atlas of normal and tumorigenic human breast tissue | Scientific Data

A single-cell Arabidopsis root atlas reveals developmental trajectories in wild-type and cell identity mutants - ScienceDirect

Longitudinal single-cell RNA-seq analysis reveals stress-promoted chemoresistance in metastatic ovarian cancer | Science Advances

Single-Cell RNA Sequencing and Assay for Transposase-Accessible Chromatin Using Sequencing Reveals Cellular and Molecular Dynamics of Aortic Aging in Mice | Arteriosclerosis, Thrombosis, and Vascular Biology

Computational approaches for interpreting scRNA‐seq data - Rostom - 2017 - FEBS Letters - Wiley Online Library
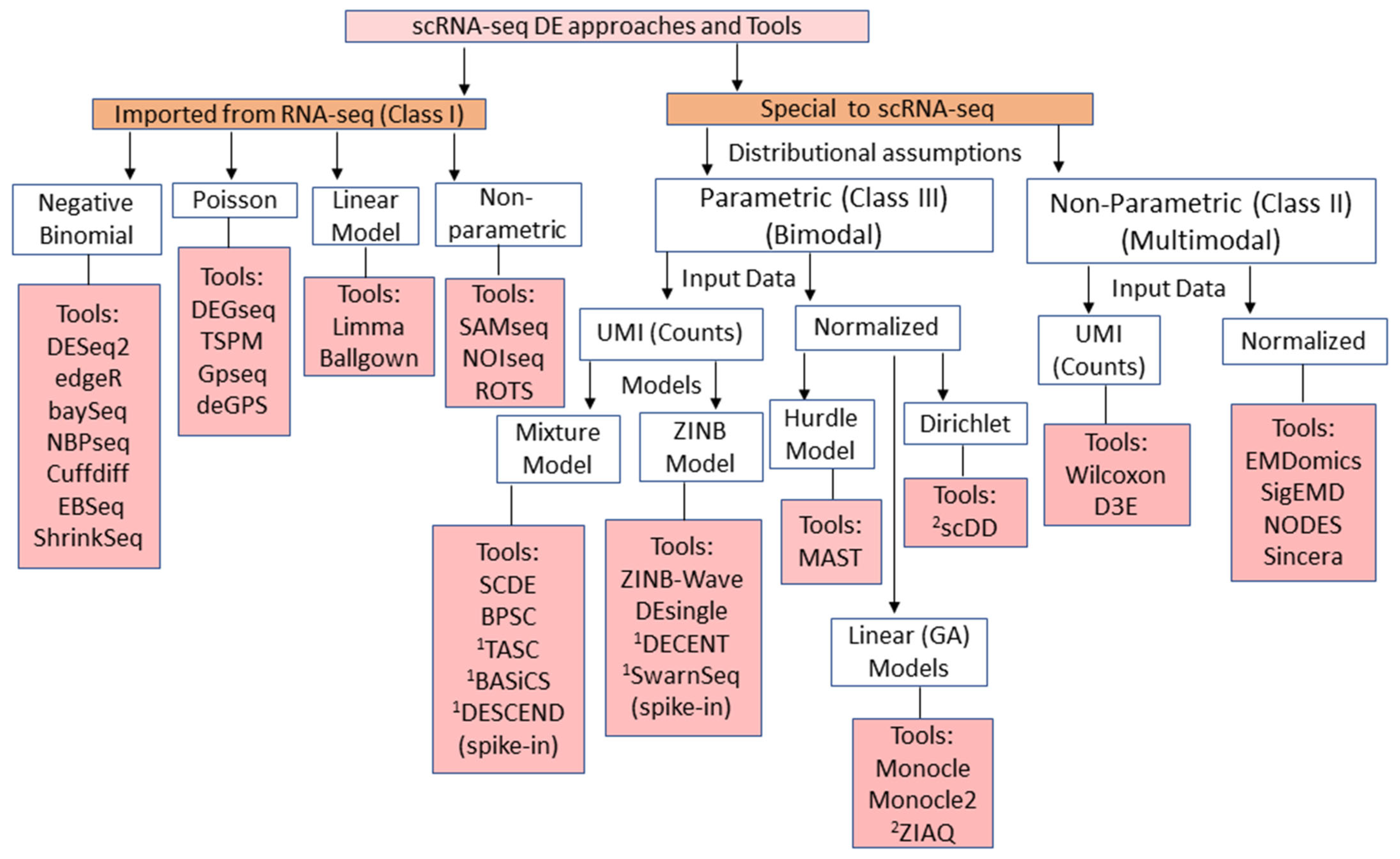
Genes | Free Full-Text | A Comprehensive Survey of Statistical Approaches for Differential Expression Analysis in Single-Cell RNA Sequencing Studies
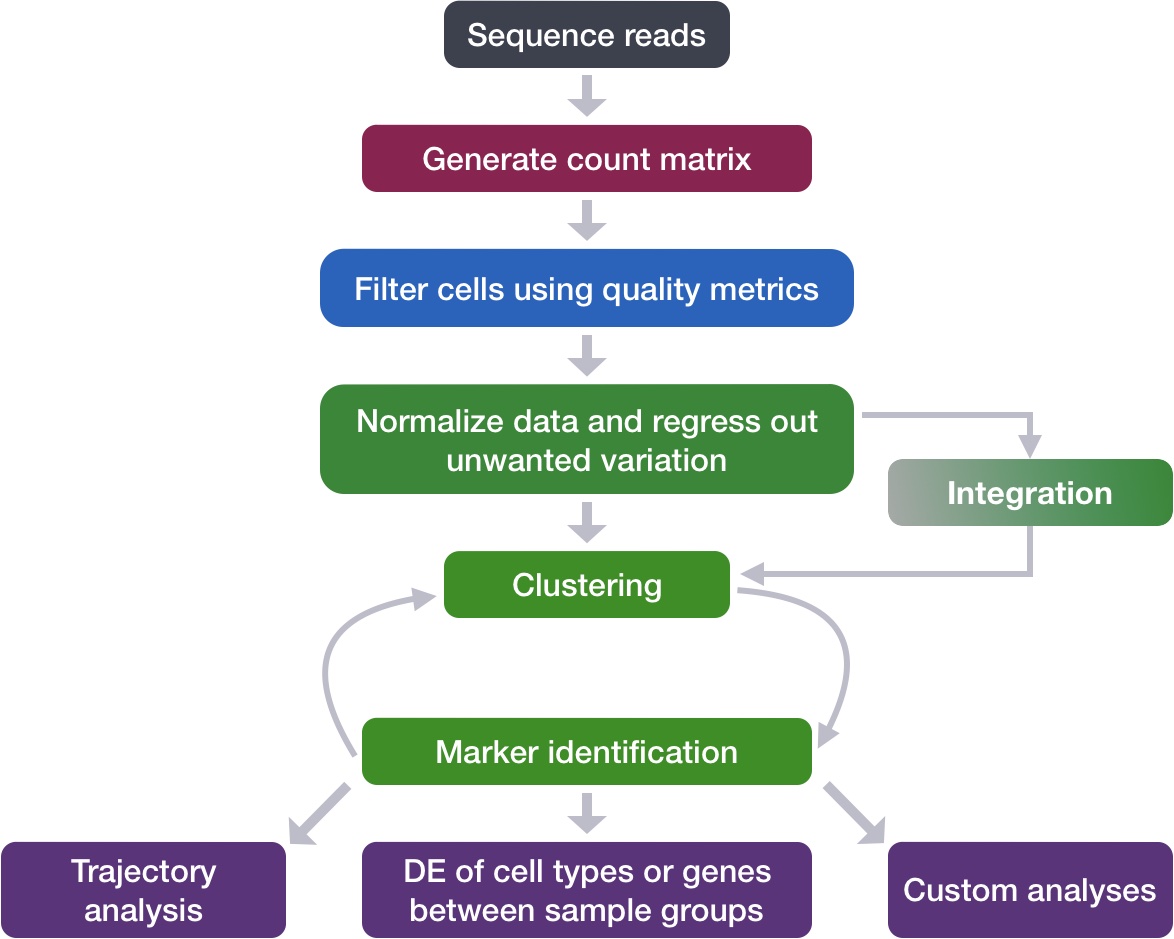
Single-cell RNA-seq: Pseudobulk differential expression analysis | Introduction to single-cell RNA-seq
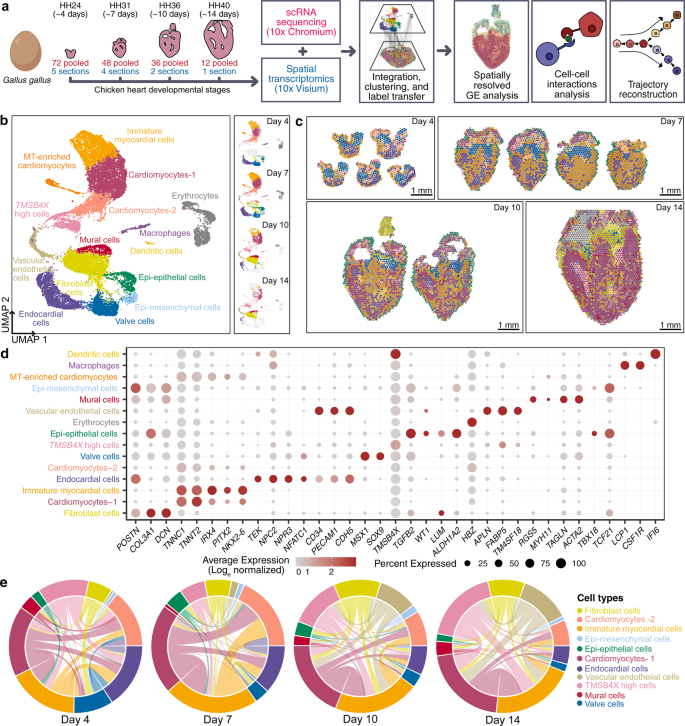
Spatiotemporal single-cell RNA sequencing of developing chicken hearts identifies interplay between cellular differentiation and morphogenesis | Nature Communications
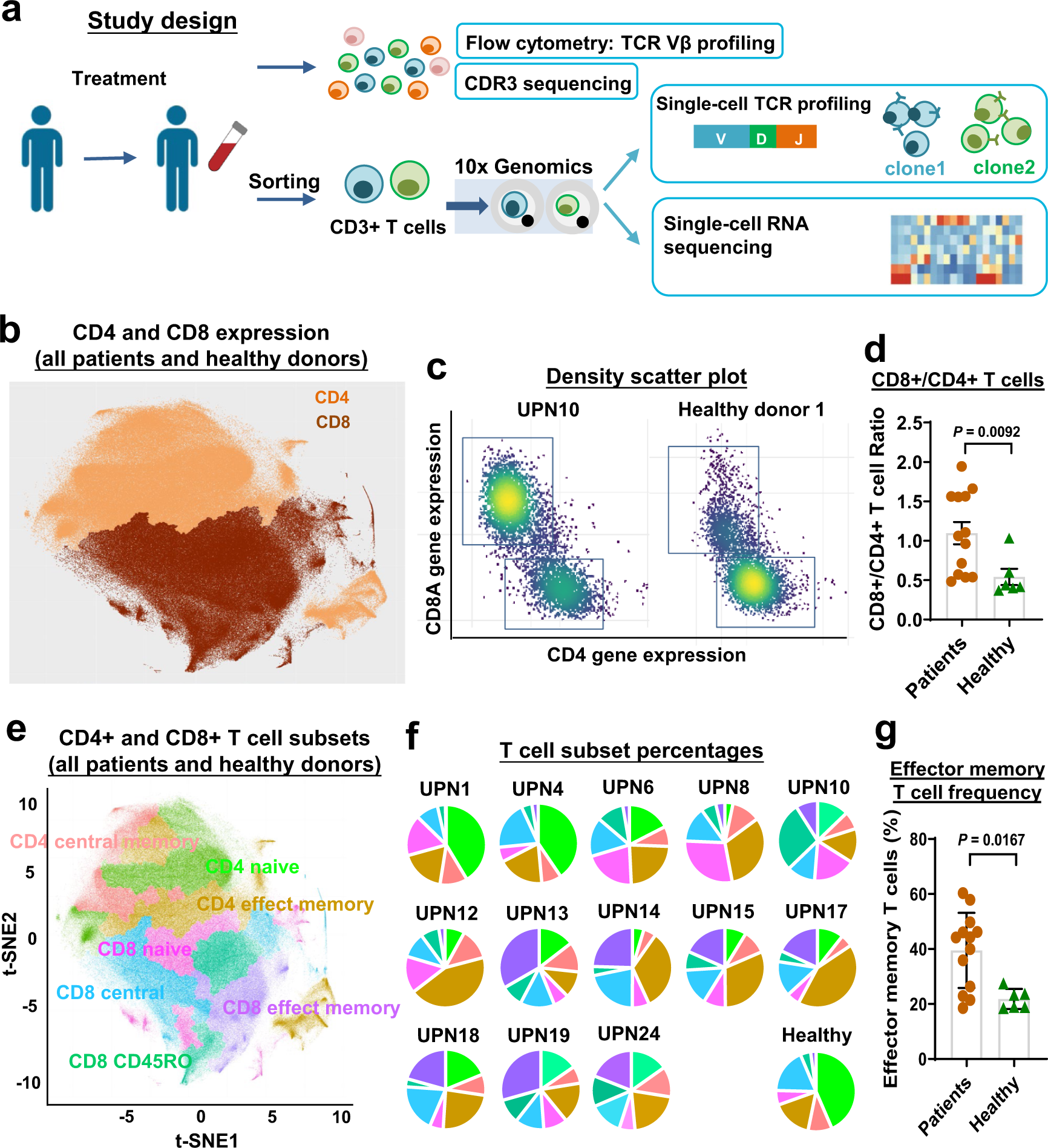
Single-cell RNA sequencing coupled to TCR profiling of large granular lymphocyte leukemia T cells | Nature Communications
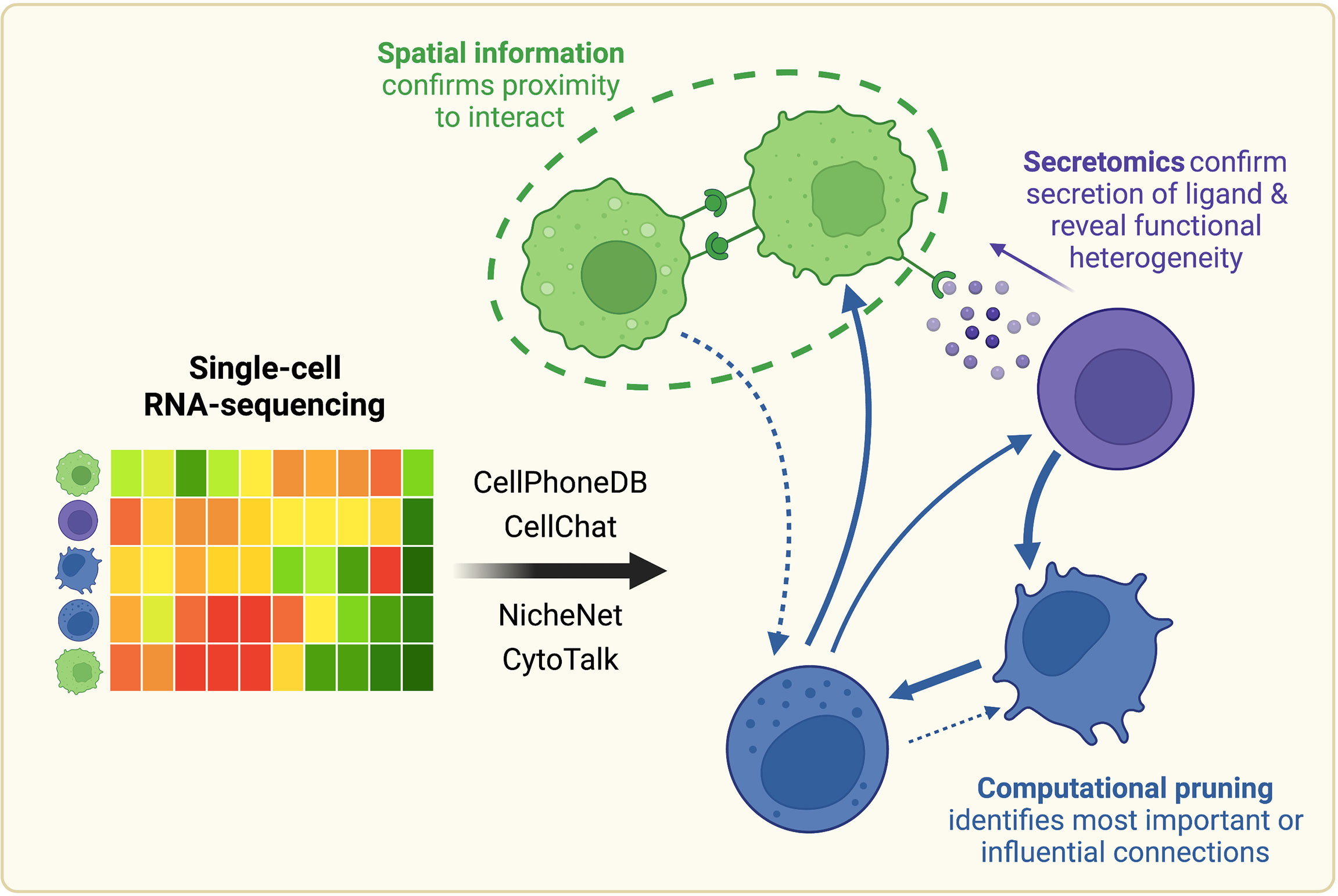
Frontiers | Mapping and Validation of scRNA-Seq-Derived Cell-Cell Communication Networks in the Tumor Microenvironment
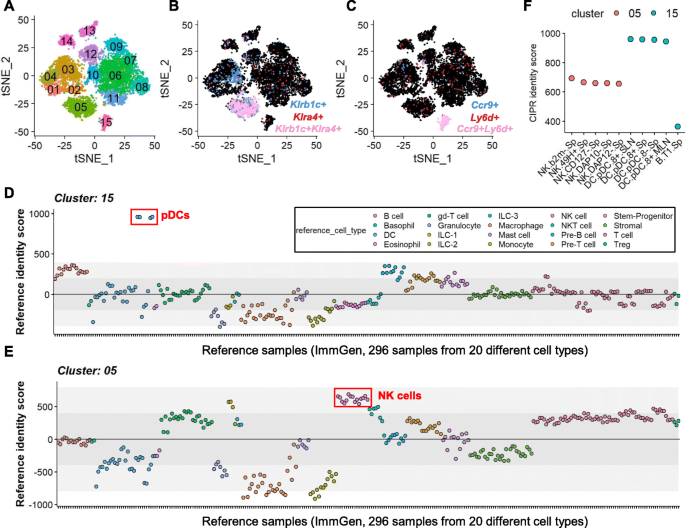
CIPR: a web-based R/shiny app and R package to annotate cell clusters in single cell RNA sequencing experiments | BMC Bioinformatics | Full Text
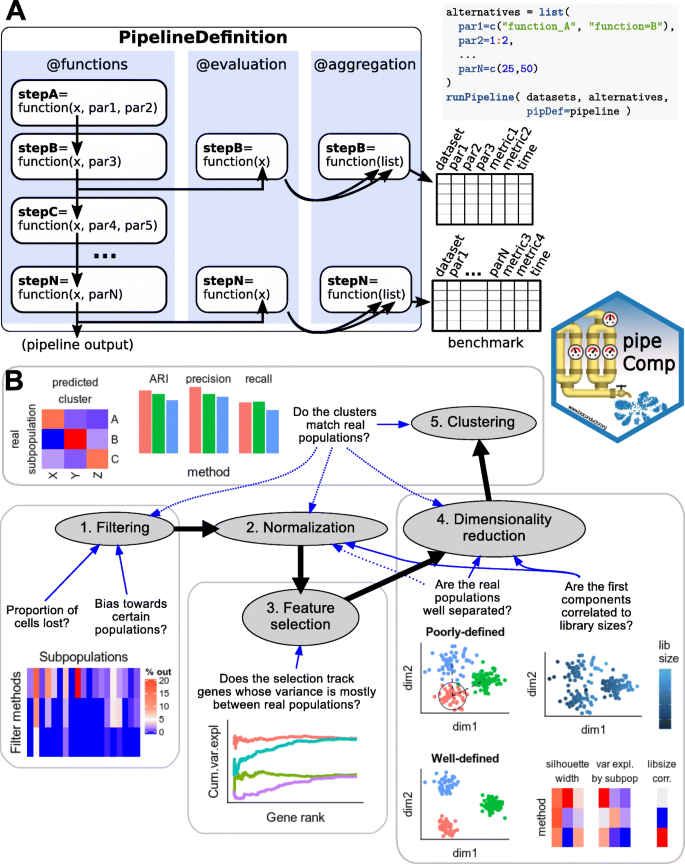
pipeComp, a general framework for the evaluation of computational pipelines, reveals performant single cell RNA-seq preprocessing tools | Genome Biology | Full Text

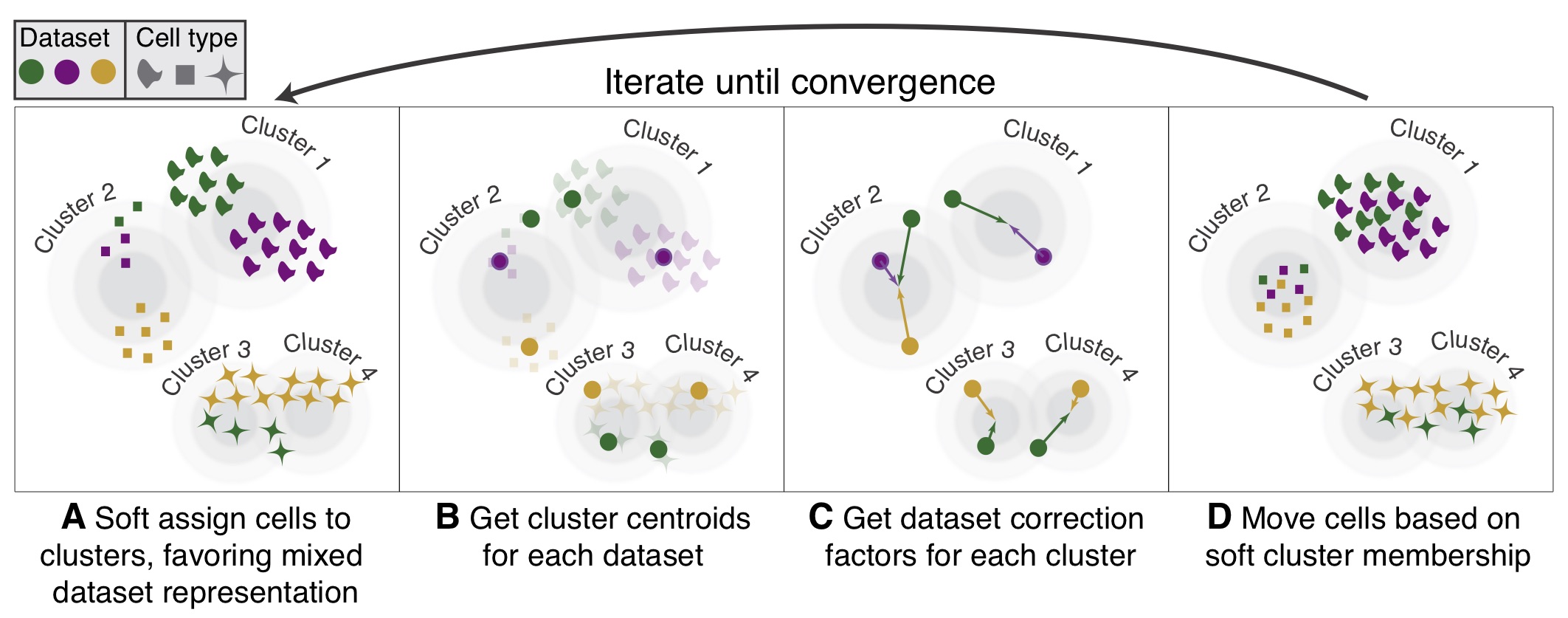


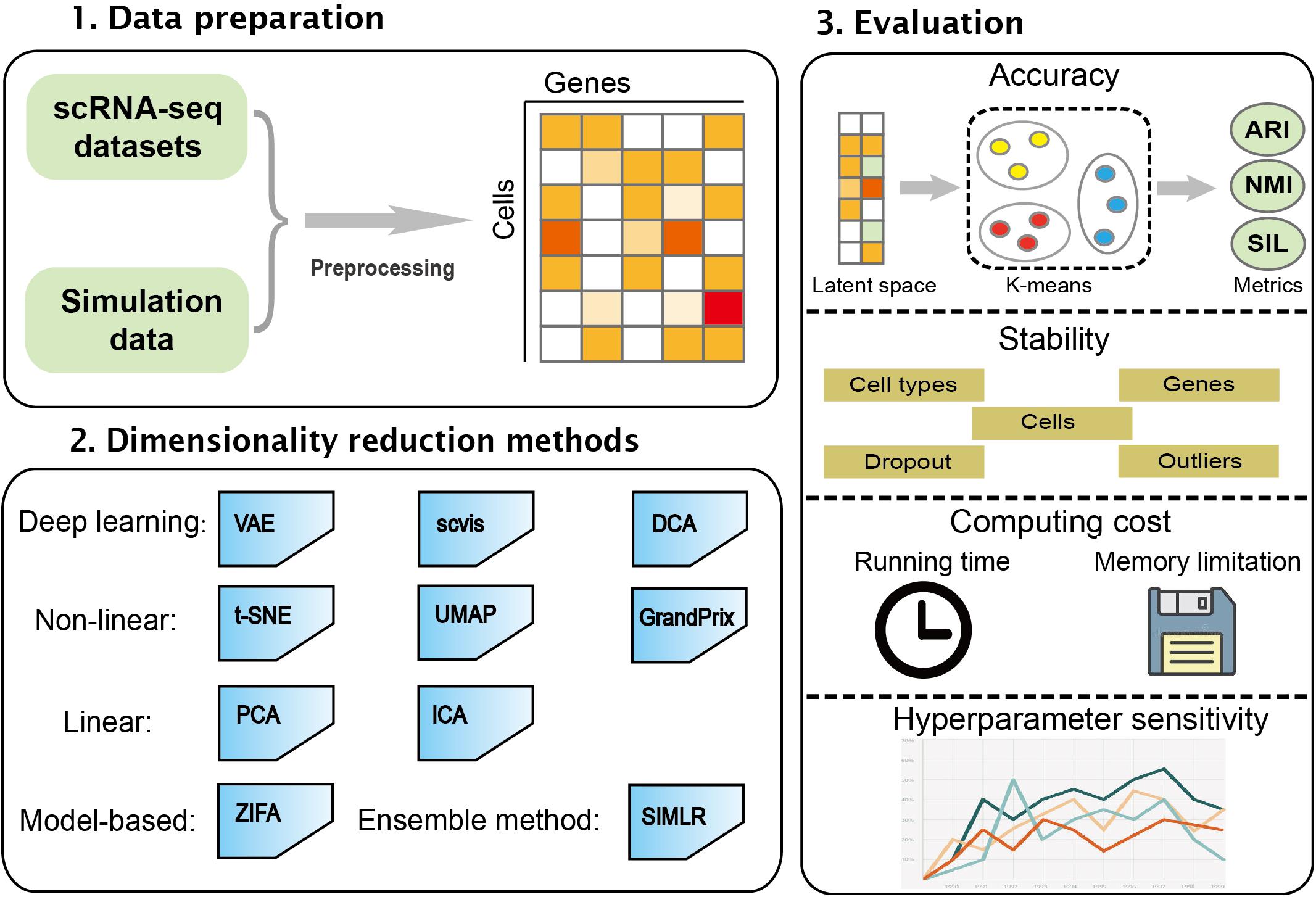



![BingleSeq: a user-friendly R package for bulk and single-cell RNA-Seq data analysis [PeerJ] BingleSeq: a user-friendly R package for bulk and single-cell RNA-Seq data analysis [PeerJ]](https://dfzljdn9uc3pi.cloudfront.net/2020/10469/1/fig-1-2x.jpg)